paper reading list
Connectome
connectome dataset
MICrONS Project
technology of connectome
connection reconsturction
connectome analysis
FT(T;X)=c∈C(X)∑(nTnc&T)2
- Visual Columns Map
A comprehensive mapping of the 31 columnar cell types in the compound eye has been achieved using a computational optimization technique based on synaptic connectivity. A manuscript describing the resource and methodology is currently in preparation. The quality of this mapping can be evaluated through specially developed visualizations available below.
connectome and gene
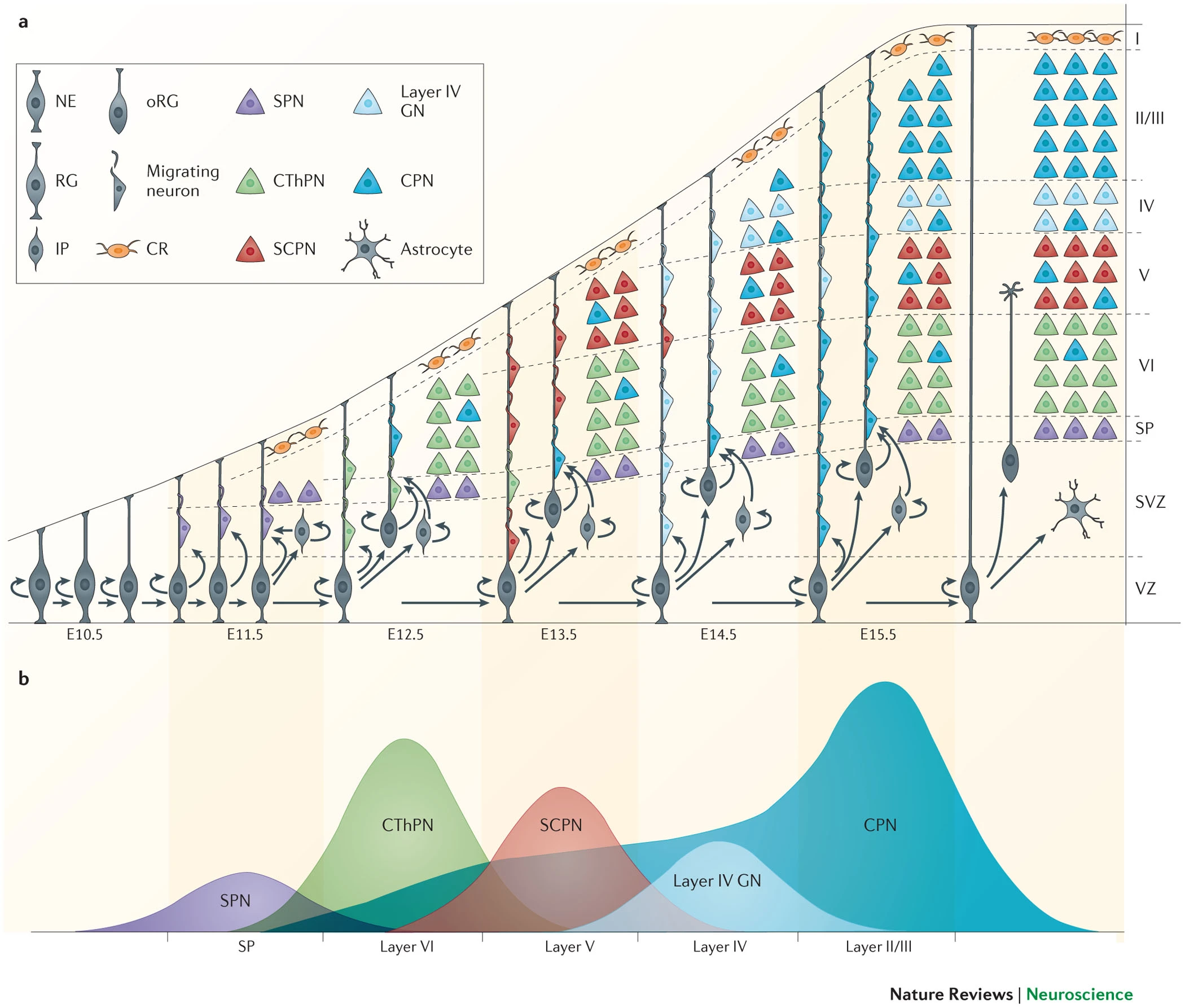
review
-
The cell biology of synapse formation SAM
Review, 2021, from Thomas C. Südhof

synaptic adhesion molecules.png
-
Towards an Understanding of Synapse Formation
Review, 2018, from Thomas C. Südhof
-
The Dynamic Synapse Review, 2013, Neuron, Daniel Choquet, Antoine Triller
-
Making Connections in the Fly Visual System
Review, 2002, Thomas R Clandinin, S.Lawrence Zipursky
-
Synaptic Connectivity and Neuronal Morphology: Two Sides of the Same Coin
-
Beyond Molecular Codes: Simple Rules to Wire Complex Brains
Review, 2015, Bassem A. Hassan, P. Robin Hiesinger
-
Chemoaffinity Revisited: Dscams, Protocadherins, and Neural Circuit Assembly
Teview, 2010, S. Lawrence Zipursky, Joshua R. Sanes
-
Architectures of neuronal circuits
reivew, 2021, Science, Liqun Luo

computational methods
experimental test
data:
other methods to reconstruct connectome
human connectome
experiment support
cell-cell communication and interaction
protein interaction
experiment support
deep learning methods
result verification
- A conserved fertilization complex bridges sperm and egg in vertebrates AlphaFold-Multimer
This approach employs structural predictions of pairwise protein-protein interactions between each bait protein and the candidate proteins. A high interface-predicted template modeling score (ipTM score) reflects high confidence in the relative positioning of the interfaces.

data mapping between transcriptome and connectome
neural type
my blog on connectivity_to_type
p(y∣Q,f)∝e−21f′Mf+y′Qf
neural activity and connectivity
morphology
-
Mapping Function Onto Neuronal Morphology
Klaus M. Stiefel, Terrence J. Sejnowski
We used a Lindenmayer-system (L-system) (Lindenmayer 1968) for the algorithmic construction of model neuron morphologies. Simulations of electrophysiological dynamics were carried out in the neuronal simulation language NEURON (version 5.7) (Hines and Carnevale 1997, 2000).
-
Neuronal contact predicts connectivity in the C. elegans brain
Peters’ rule explains synaptic specificity throughout the C. elegans nervous system
-
Development of Dendritic Form and Function
Review paper, 2015, Julie L. Lefebvre, Joshua R. Sanes, and Jeremy N. Kay
-
One Rule to Grow Them All: A General Theory of Neuronal Branching and Its Practical Application
2010
-
Molecular mechanisms of dendrite morphogenesis
Review, 2012, Jyothi Arikkath
-
Automated analysis of neuronal morphology, synapse number and synaptic recruitment
-
L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies
-
TreeMoCo: Contrastive Neuron Morphology Representation Learning
-
Principles of branch dynamics governing shape characteristics of cerebellar Purkinje cell dendrites
Growing dendrites are retracted or stalled by contacts with other dendrites.
morphology data
NeuroMorpho.Org is a centrally curated inventorys of digitally reconstructed neurons and glia
maximum weighted independent set (MWIS)
Graph compression or sparsification
Maximum Dispersion
Maximum Diversity Problem (MDP)
connectome visualization
human connectome
gene data
gene data for Drosophila
analysis
logical gene for connectome
gene regulatory network
Gene pairs bulkCOPs ¬ bulkCOPs scCOPs 3133245¬ scCOPs 2993232603
review paper
sota models
application
gene normalization
functional and structure conenctome
-
Integrating multimodal and multiscale connectivity blueprints of the human cerebral cortex in health and disease
-
The cell-type underpinnings of the human functional cortical connectome
-
Complex brain networks: graph theoretical analysis of structural and functional systems
Ed Bullmore & Olaf Sporns (2009).
Nature reviews neuroscience
-
Engel, A. K., Gerloff, C., Hilgetag, C. C., & Nolte, G. (2013). Intrinsic coupling modes: multiscale interactions in ongoing brain activity. Neuron, 80(4), 867-886.
-
Biswal, B., F. Zerrin Yetkin, Victor M. Haughton, James S. Hyde (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magnetic Resonance in Medicine 34(4): 537-541.
-
Markov NT, et al. (2014) A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb Cortex 24:17–36.
-
Donahue, C.J., Sotiropoulos, S.N., Jbabdi, S., Hernandez-Fernandez, M., Behrens, T.E., Dyrby, T.B., Coalson, T., Kennedy, H., Knoblauch, K., Van Essen, D.C., Glasser, M.F. (2016). Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J. Neurosci. 36, 6758–6770. doi:10.1523/JNEUROSCI.0493-16.2016.
-
Honey, C., Thivierge JP, Sporns O (2010). Can structure predict function in the human brain? Neuroimage 52(3): 766-776.
-
Deco, G., McIntosh, A. R., Shen, K., Hutchison, R. M., Menon, R. S., Everling, S., Hagmann, P., & Jirsa, V. K. (2014). Identification of optimal structural connectivity using functional connectivity and neural modeling. The Journal of neuroscience : the official journal of the Society for Neuroscience, 34(23), 7910–7916.
-
Dawson, DA, Cha K, Lewis LB, Mendola JD, Shmuel A (2013). "Evaluation and calibration of functional network modeling methods based on known anatomical connections." Neuroimage 67: 331-343.
-
Predicting brain structural network using functional connectivity. Medical image analysis, 79, 102463. https://doi.org/10.1016/j.media.2022.102463.
-
Predicting brain structural network using functional connectivityj.media
multi-GCN based GAN, AD
-
Prediction of brain sex from EEG: using large-scale heterogeneous dataset for developing a highly accurate and interpretable ML model
-
Topographic Axes of Wiring Space Converge to Genetic Topography in Shaping the Human Cortical Layout
-
A practical guide to linking brain-wide gene expression and neuroimaging data
other connectome model:
Reinforcement learning (RL)
graph
Link prediction
https://paperswithcode.com/task/link-prediction
review
Graph Embedding
review
| Dataset | Nodes | Node Types | Edges | Edge Types | Target |
|---------|-------|------------|-------|------------|--------|
| Amazon | 10,099| 1 | 148,659| 2 | product-product |
| LastFM | 20,612| 3 | 141,521| 3 | user-artist |
| PubMed | 63,109| 4 | 244,986| 10 | disease-disease |
disease gene
graph and connectome
BrainGNN: Interpretable Brain Graph Neural Network for fMRI Analysis
https://www.sciencedirect.com/science/article/abs/pii/S1361841521002784
Graph Neural Networks for Brain Graph Learning: A Survey
https://arxiv.org/abs/2406.02594
2024
dynamics system & Ripple
connectome inspired neural network
html connectome inspired neural network
-A model of a CA3 hippocampal pyramidal neuron incorporating voltage-clamp data on intrinsic conductances
R. D. Traub,R. K. Wong,R. Miles, andH. Michelson, 1991
Soma compartments and neurite compartments
Spiking neural network
single neuron dynamics
U˙=−gLU+gD(Vw−U)+IUsom IUsom (t)=gE(t)(EE−U)+gI(t)(EI−U).
dtdui(x)=−τxui(x)−EL+Cxgxf(ui(d))+cxK(t−t^i(s))+Ii(x)+wi(x)dtdwi(x)=−wi(x)/τw(x)+aw(x)(ui(x)−EL)/τw(x)+bw(x)Si(x),
-
Impact of dendritic size and dendritic topology on burst firing in pyramidal cells
E. van, A. J. Ronald and A. Ooyen,PLoS Comput. Biol., 2010
Morphologically simplified cells, the set of 23 neurons
-
Distinguishing theoretical synaptic potentials computed for different soma-dendritic distributions of synaptic input
W. Rall, J. Neurophysiol., 1967
-
Models of Neocortical Layer 5b Pyramidal Cells Capturing a Wide Range of Dendritic and Perisomatic Active Properties
Etay Hay ,Sean Hill,Felix Schürmann,Henry Markram,Idan Segev

HH model
Hodgkin & Huxley 1952
LIF
GLIF model
synapse
-
A role for NMDA-receptor channels in working memory
Xiao-Jing Wang,1998
-
Distinguishing theoretical synaptic potentials computed for different soma-dendritic distributions of synaptic input Wilfrid Rall, 1967
-
The Contribution of AMPA and NMDA Receptors to Persistent Firing in the Dorsolateral Prefrontal Cortex in Working Memory J Neurosci, 2020
-Synaptic AMPA Receptor Plasticity and Behavior neuron, review, 2009
-
Dendritic computations captured by an effective point neuron model
Songting Li, Nan Liu, Xiaohui Zhang, David W. McLaughlin, Douglas Zhou, and David Cai, 2019, PNAS
We derive an effective point neuron model, which incorporates an additional synaptic integration current arising from the nonlinear interaction between synaptic currents across spatial dendrites.
We then derive a form of synaptic integration current within the point neuron framework to capture dendritic effects.
hLN
experiment support
multiple recent studies indicate that dendritic activity in vivo is much less compartmentalized than what was previously hypothesized (Xu et al., 2012, Beaulieu-Laroche et al., 2019, Francioni et al., 2019, Kerlin et al., 2019, Voigts and Harnett, 2019).
The first demonstrated that somatic voltage fluctuations under an in vivo-like synaptic regime are well described by a global dendritic nonlinearity that can capture up to 90% of explained variance (Ujfalussy et al., 2018).
Michalis Pagkalos, Spyridon Chavlis & Panayiota Poirazi, 2023, Nature Communications
Tool for neuron dynamics simulation (like brian2, spikingjelly)
review
temporal inhibition
timing-dependent inhibition
modeling:
other
olfactory system
mean field
NeuroAI
Connectome & ANN
connectome inspired neural network
html connectome inspired neural network
geometry
Brain simulation / activity prediction
model
Network analysis
BOOK Fundamentals of Brain Network Analysis
BOOK Chapter 3 - Connectivity Matrices and Brain Graphs
Review Neural Networks With Motivation
synapse plasticity
STDP
nearest neighbor STDP (Izhikevich, 2003)
BTSP
neural data recording
whole brain imaging
drosophila data
other data
cell tracking
wormid
neural data analysis & modeling
reivew
high dimensional data
dimensional reduction
manifold reconstriction
activity analysis
head direction
encoding and decoding
deep learning method
fundation model
diffusion model/ visual Stimulus
behavior
oscilation
place cell and grid cell
vision
drosophila
others
ML
hippocamus
gene
cognition
attention
learning and memory
decision making
DS & OR
Quadratic assignment problem
similarity matrix
-
Euclidean Distance Matrices: Essential Theory, Algorithms and Applications
-
Representational similarity analysis – connecting the branches of systems neuroscience
-
Euclidean Distance Matrices: Essential Theory, Algorithms and Applications
-
CCA Kernel and nonlinear canonical correlation analysis
-
Similarity of Neural Network Representations Revisited
Simon Kornblith, Mohammad Norouzi, Honglak Lee, Geoffrey Hinton
-
Representational dissimilarity metric spaces for stochastic neural networks
Lyndon R. Duong, Jingyang Zhou, Josue Nassar, Jules Berman, Jeroen Olieslagers, Alex H. Williams
ML
NAS
CNN kernel
Clustering convolutional kernels
sparse decoding
UFLDL Tutorial
others
wiki
TODO



